I need to look through a certain cell line lineage for all top essential genes, for future CRISPR for potential slicing and silencing. For instance, I need all head and neck cell lines, and I’m looking for essential genes that overlap amongst them. I understand it is possible to manualy go through all 95 cell lines in that lineage, but is there any way DepMap has a built in utility for that?
Thank you.
When you say “essential genes” I’m assuming you mean essential in that line, but excluding genes which are essential in all lines, correct?
Regardless, I’m thinking about how I’d answer that question, and we don’t have a way to answer that question exactly the way you framed it (for each line, what is the set of essential genes, and then what is the overlap across sets) however, have the ability to aggregate across lines by taking the mean.
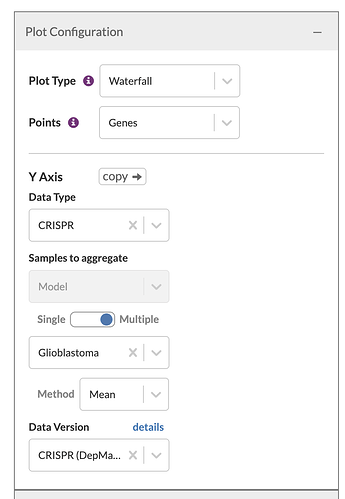
Concretely, I can say I want to see a plot of the mean gene effect for those lines in a collection of lines (I’ve chosen Glioblastoma because I had that set defined already) and I want to see the mean per gene:
I can also say that I want to break out the common essential genes because those aren’t specific to the context:
Now I can see which genes, on average, are most lethal in the lines for this disease type.
Thanks,
Phil