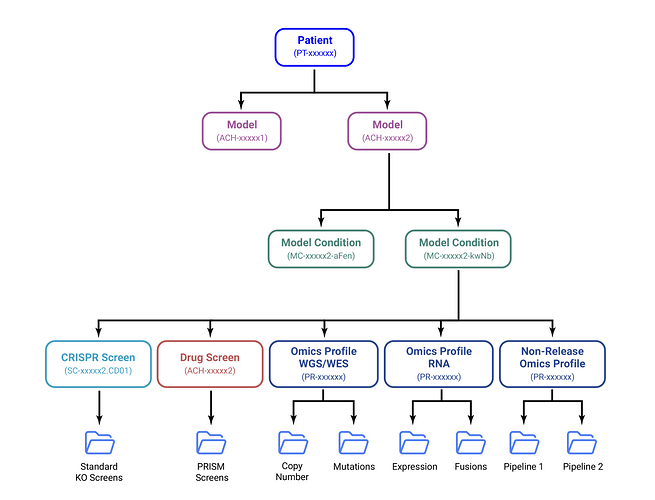
DepMap Data Structure
Depmap Releases utilize a multi-level data hierarchy to accommodate richer types of data. Each element in the hierarchy is indexed by a unique ID and defined by a set of tables in the data release.
Because there can be multiple conditions for each model, mapping files are available to help you connect IDs and better understand which data are available across our portal tools.
You can now view data for any condition for DepMap Release data in Data Explorer. However, please note that we do set a default condition for data shown.
Some collaborator datasets may follow the same structure as DepMap Release data, but are considered independent. This data may be available to you in Data Explorer as additional files.
-
At the top of the hierarchy is Patient.
-
Models are a collection of cells derived from a single biopsy of the Patient. Each Patient can have one or more derived Models.
-
CRISPR and sequencing data are generated from a Model Condition. Each CRISPR Screen receives a Screen ID. Each sequencing datatype (e.g. wgs, rna, wes, etc.) receives an Omics Profile ID. Non-release Omics datasets (OLINK, ATAC-Seq) also receive an Omics Profile ID, but are not considered part of the bi-annual DepMap Release Dataset.
-
Although data is generated from Model Condition, DepMap Release data are indexed at two principal levels: Models and Screens/Profiles.
For more information, head to our data hub here.